Postdoctoral research fellow
Nephrology
Toronto General Research Institute, University Health Network
Protein and miRNA expression distinguish DSA+ kidney transplant recipients with and without antibody-mediated rejection
Kieran Manion1, Trirupa Chakraborty2, Sergi Clotet-Freixas1, Sofia Farkona1, Alex Boshart1,3, Antoninus Soosaipillai4, Rohan John1, Jishnu Das2, Ana Konvalinka1,3,5.
1Toronto General Research Institute, University Health Network, Toronto, ON, Canada; 2University of Pittsburgh School of Medicine, University of Pittsburgh, Pittsburgh, PA, United States; 3Institute of Medical Science, University of Toronto, Toronto, ON, Canada; 4Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, ON, Canada; 5Ajmera Family Transplant Centre, University Health Network, Toronto, ON, Canada
Introduction: As of 2020, over 41,000 Canadians are living with end-stage kidney disease, where the kidneys no longer have sufficient function to maintain body homeostasis. While transplantation is the best treatment for end stage kidney disease, 50% of grafts fail within 10 years, due primarily to antibody-mediated rejection (ABMR), where recipient donor-specific antibodies (DSA) trigger immune-driven tissue injury. There are no effective treatments for ABMR and predicting its onset is complex, as 30-60% of DSA+ transplant patients do not develop rejection. The purpose of our study is to identify factors that regulate kidney protein expression in DSA+ ABMR kidney transplant recipients compared to DSA+ recipients without rejection.
Methods: Laser capture microdissection was used to extract glomeruli and tubulointerstitium from kidney biopsies from DSA+ kidney transplant recipients with ABMR (n=14) or no rejection (NR; n=16) (cohort 1). Protein from these tissues was subjected to liquid chromatography mass spectrometry on Q-Exactive HFX and results were analyzed using MaxQuant and Perseus software. Significantly differentially expressed proteins (t-test, p<0.05) were mapped to signalling pathways via pathDIP and Reactome databases (FDR q value<0.05), and associated miRNAs were identified via the miRDIP database. miRNA was measured (HTG Molecular Diagnostics) in the sera of a second DSA+ patient cohort (n=40) and assessed for association with ABMR via LASSO and Essential Regression machine learning.
Results: We identified 184 proteins in glomeruli and 170 in the tubulointerstitium (FDR<1%) that were significantly differentially expressed between DSA+ ABMR and NR patients in cohort 1, with 61% and 73% upregulated in ABMR, respectively. pathDIP and Reactome analyses revealed that these proteins mapped significantly to pathways involved in the immune and cellular stimuli responses (Fig. 1A), particularly antigen processing (q= 6.49e-12) and oxidative stress (q= 1.75e-08). LASSO analysis of miRNA measured in the serum of a second cohort of DSA+ patients revealed 12 miRNA that significantly discriminated between patients with or without ABMR (Fig. 1B). Subsequent miRDIP analysis found that all 12 of the identified miRNAs target the genes encoding kidney proteins differentially expressed in cohort 1.
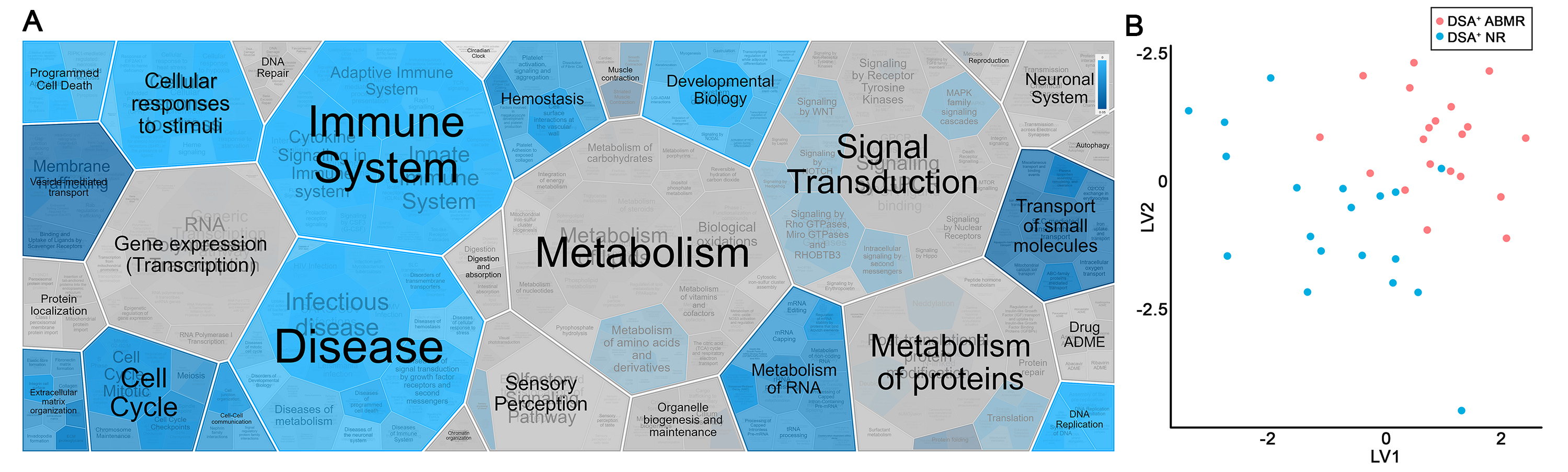
Conclusions: These preliminary results suggest that ABMR in DSA+ patients is strongly linked to dysregulated immune and cellular stress responses and that this dysregulation manifests in multiple, interconnected cellular products. These findings will ultimately help us identify targets for the development of novel therapeutics for kidney transplant recipients.
References:
[1] CORR ESKD Tables and Figures: 2011 to 2020 Data. CIHI 2022.
[2] Wolfe R, et al. N Engl J Med 1999. 341: 1725-1730.
[3] Rana A, et al. JAMA Surg 2015. 150: 252.
[4] Stegall M, et al. JASN 2015. 26: 20–29.
[5] Sellares J, et al. Am J Transplant 2012. 12: 388-399.
[6] Gaston R, et al. Transplantation 2010. 90: 68-74.
[7] Valenzuela N, et al. Am J Transplant 2015. 15: 1502-1518.
[8] Hirohashi T, et al. Am J Transplant 2012. 12: 313-321.
[9] Lefaucheur C, et al. JASN 2016. 27: 293–304.
[10] Clotet-Freixas S, et al. JASN 2020. 31: 2705–2724.
Lectures by Kieran Manion
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Tue-02 16:15 - 17:20 |
Abstracts Session 5 | Protein and miRNA expression distinguish DSA+ kidney transplant recipients with and without antibody-mediated rejection | Grand Georgian |
