David A Hildeman, United States
Professor
Immunobiology
Cincinnati Children's Hospital Medical Center
Tracking gene expression and clonal expansion of CD8+ T cells in the biopsies and urine of patients undergoing kidney transplant rejection
David Hildeman2, Tiffany Shi2, Ashley Burg2, J. Tim Caldwell5, Krishna Roskin4, Adele Shields3, Rita Alloway3, E. Steve Woodle1.
1Surgery, University of Cincinnati, Cincinnati, OH, United States; 2Immunobiology, Cincinnati Children's Hospital, Cincinnati, OH, United States; 3Nephrology, University of Cincinnati, Cincinnati, OH, United States; 4Bioinformatics, Cincinnati Children's Hospital, Cincinnati, OH, United States; 5Nephrology and Hypertension, Cincinnati Children's Hospital, Cincinnati, OH, United States
Introduction: Although CD8+ T cells are the major drivers of acute cellular rejection (ACR) across multiple organs, little is known about their biology and persistence during rejection.
Methods: We performed single cell RNA + TCR-CDR3αβ sequencing (scRNAseq, scTCRseq) of renal allograft biopsies from 10 patients undergoing ACR while on 3 different types of maintenance immunosuppression (tacrolimus, iscalimab (anti-CD40 mAb), or belatacept (CTLA4-Ig)). In addition, we performed temporal scRNAseq/scTCRseq on biopsies and urine from 3 patients undergoing anti-rejection therapy.
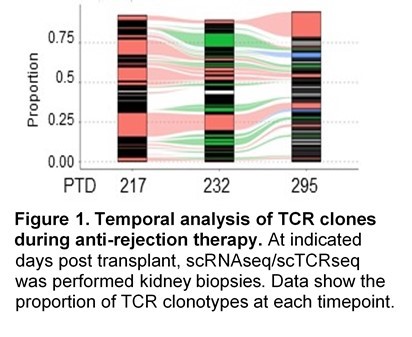
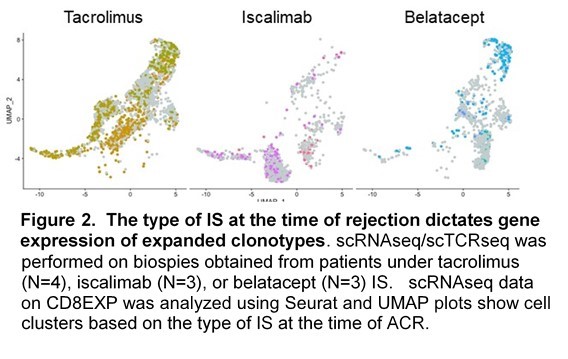
Results: We observed a stunning degree of TCR restriction (~20 expanded CD8+ clonotypes [CD8EXP]/patient). This clonal restriction was not due to sampling bias, as clonotypes observed at initial ACR were observed in subsequent persistent rejection biopsies (Fig. 1) as well as in paired urine samples. Further, unexpanded clones at one timepoint were rarely observed as expanded at a later timepoint. Transcriptomic analysis of expanded clonotypes (CD8EXP) demonstrated a variety of cellular states/phenotypes, including circulating memory, resident memory, and varying degrees of exhaustion. Intriguingly, a resident memory phenotype was found in the CD8EXP in the 2 patients in this cohort with persistent rejection and eventual graft loss. In contrast, rejection resolution was associated with a near elimination of CD8EXP in the biopsy. Next, we found unique DEG signatures of the CD8EXP for each IS therapy, suggesting that IS therapy determines the gene expression of CD8EXP (Fig.2). Our data suggest that these transcriptomic profiles may assist in the prediction of responsiveness to particular anti-rejection therapies. Finally, we found that CD8EXP that persist after anti-rejection therapy often change their transcriptomic phenotype, which may contribute to their lack of responsiveness to therapy.
Conclusion: Together our data show that ACR is characterized by: 1) a remarkably limited number of distinct CD8EXP, 2) CD8EXP that persist in ongoing rejection and disappear with rejection resolution, 3) the presence of clonally identical CD8EXP in the urine and kidney biopsies from the same patients, 4) a persistence of CD8EXP with a resident memory phenotype which is associated with later graft loss, 5) distinct transcriptional profiles of CD8EXP depending on the type of IS therapy, 6) transcriptomic adaptation of CD8EXP in response to anti-rejection therapy. Combined, our data provide several novel insights into the biology of ACR and strongly argue for a personalized approach to understanding ACR to guide anti-rejection therapies.
The authors acknowledge funding from Novartis and from NIH R21AI142264.
Lectures by David A Hildeman
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Sun-30 16:40 - 18:00 |
Abstracts Session 1 | Tracking gene expression and clonal expansion of CD8+ T cells in the biopsies and urine of patients undergoing kidney transplant rejection | Grand Georgian |
