Khodor I Abou-Daya, United States
Assistant Professor of Surgery
Surgery
Thomas E. Starzl Transplantation Institute
The epigenetic and transcriptional landscape of monocyte memory formation in response to allogeneic non-self
Jason Ossart1, Neda Feizi1, Matthieu Heitz2, Sarah Masri2, Hehua Dai1, Amanda Williams1, Aravind Cherukuri1, Fadi Lakkis1, Geoffrey Schiebinger2, Martin Oberbarnscheidt1, Khodor Abou-Daya1.
1Thomas E. Starzl Transplantation Institute, Department of Surgery, University of Pittsburgh, Pittsburgh, PA, United States; 2Department of Mathematics, University of British Columbia, Vancouver, BC, Canada
Introduction: Murine monocytes require A-type paired immunoglobulin-like receptors (PIR-As) to specifically recognize and acquire memory to major histocompatibility complex I antigens. Beyond the need for PIR-As, little is known about the mechanisms of specific innate immune memory. In this study, we aim to explore possible mechanisms of monocyte memory by investigating the epigenetic and transcriptional changes that specifically occur after allogeneic stimulation in splenic monocytes.
Methods: B6.RAG-/-γc-/- (BRG) mice were immunized by intraperitoneal injection of 20 million Balb/c irradiated splenocytes (allo). Splenic monocytes were FACS sorted at 0, 3, 7 and 28 days post-immunization. Simultaneous snRNAseq and snATACseq was then performed. To identify the changes that are specific to allo-stimulation and related to monocyte memory, monocytes were also sorted and sequenced from BRG injected with irradiated B6 splenocytes (syn) and allo-immunized BRG PIR-A-/- mice at day 7 after injection.
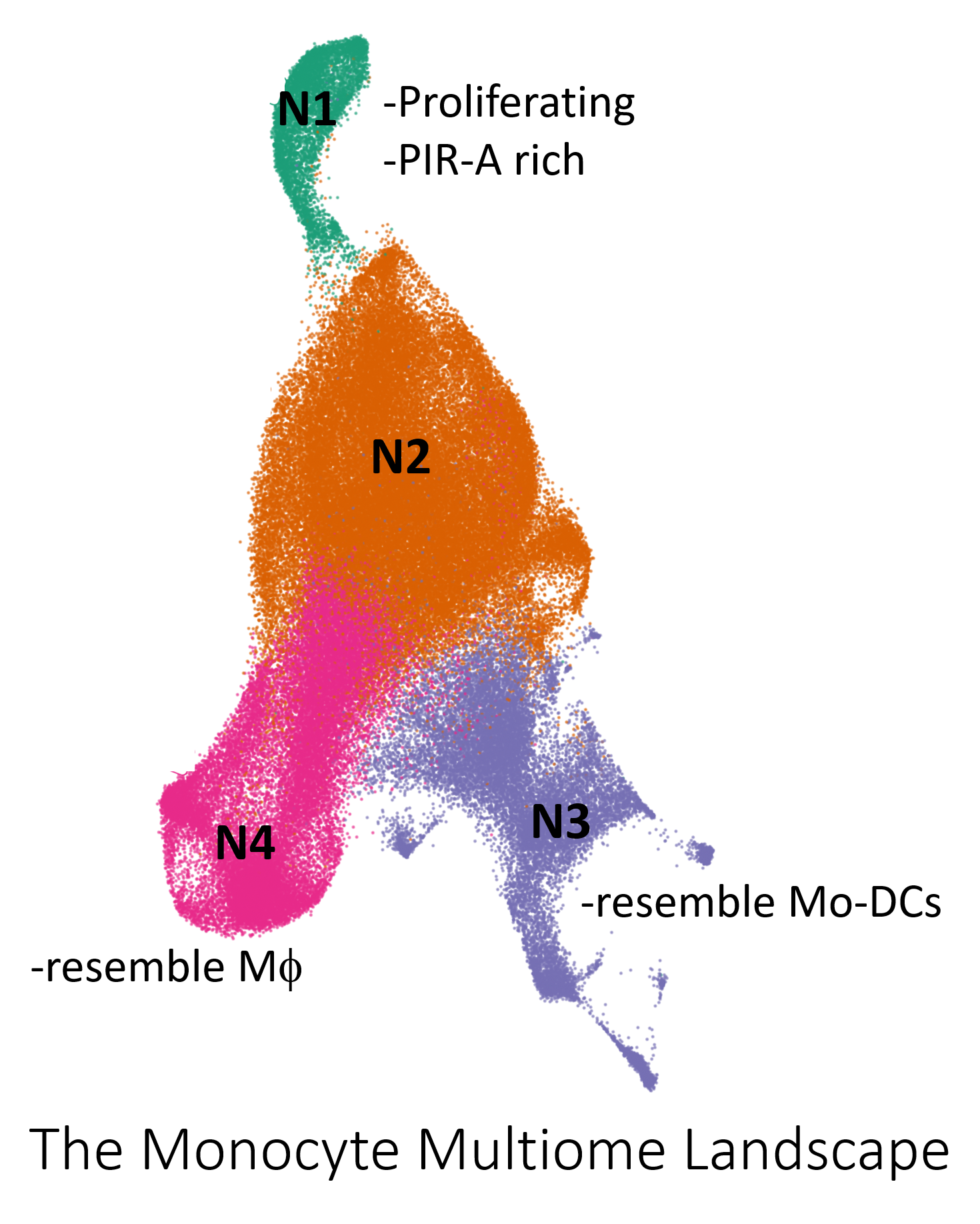
Results: Weighted nearest neighbor UMAP represented 4 visually distinct cell neighborhoods (N) (Fig. 1). N1 and N2 increased in abundance after allo-stimulation. Differential gene expression analysis revealed that N1 highly expressed genes encoding cell cycle proteins, Ly6C, and PIR-A. Monocytes in N2 from syn vs allo immunized groups were segregated. Clustering mainly resulted in the division of N2 into 3 clusters (C). C3 and N1 were more abundant in allo-immunized BRG and had a 3.5 and 2.4 fold reduction in allo-immunized BRG PIR-A-/- mice respectively. Flow cytometric analysis with EDU pulse-chase confirmed the patterns seen in the sequencing data. Multi-omic real time trajectory inference using Waddington OT supported that N1 is the starting state. Multiomic pseudotime trajectory inference using MultiVelo revealed an influx of cells from N1 to N2 7 days after allostimulation that is absent in allo-immunized BRG PIR-A-/- mice. Both real time and pseudotime trajectory inference methods suggested that the differentiation pathway connects N1 to N2 then splits to N3 and N4. N3 and N4 had a profile consistent with differentiation to macrophages and mo-DCs respectively. Gene expression and pathway analyses uncovered unique transcription factors, genes and gene networks associated with immunological memory in N1. Dictys, a multiomic dynamic gene regulatory network inference method, pinpointed active transcription factors related to the influx of cells from N1 to N2 after allostimulation.
Conclusion: Splenic monocytes are heterogenous and contain a subset that responds specifically to allo-stimulation with unique transcriptional and epigenetic features that are consistent with immunological memory formation.
University of Pittsburgh Stuart K. Patrick Grant for Transplant Innovation (Khodor Abou-Daya). NIH-NIAID R01 AI099465 (Fadi Lakkis). NIH-NIAID R01 AI145881 (Martin Oberbarnscheidt). NIH-NIAID R01 AI72973 (Fadi Lakkis). American Society of Transplantation Career Transition Grant (Khodor Abou-Daya). American Society of Transplantation Fellowship Research Grant (Neda Feizi).
Lectures by Khodor I Abou-Daya
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Wed-03 11:00 - 12:35 |
New perspectives in innate/ILC immunity and tissue-resident leukocytes | Innate allorecognition and memory | Grand Georgian |
|
Sun-30 16:40 - 18:00 |
Abstracts Session 1 | The epigenetic and transcriptional landscape of monocyte memory formation in response to allogeneic non-self | Grand Georgian |
